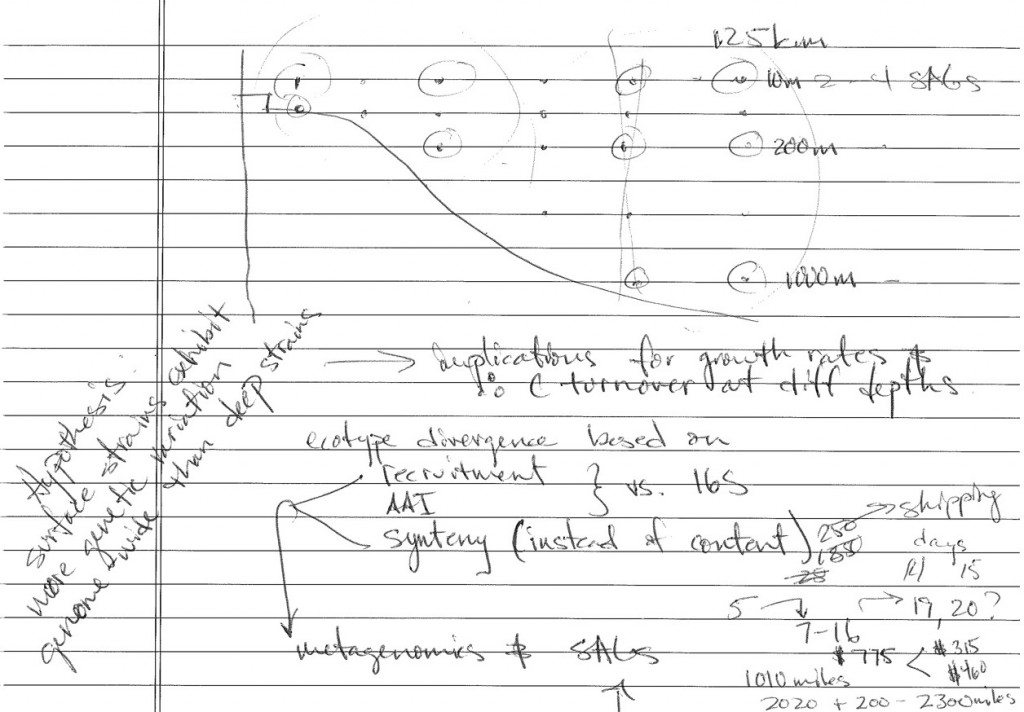
I know you will have read the scientist’s statements by now (wink wink, nudge nudge), and thus be completely up to date on what we are all trying to do and how. Ben Temperton and I are utilizing the same samples to accomplish different goals. The bulk of our work from metagenomics and single-cell genomics will come much later, after we sequence the DNA from our filters and our isolated cells. Our ship time is thankfully pretty straightforward, by design. While the ship has traveled all around the Southern California bight to serve the varied needs of the science team, we are taking samples specifically along a coastal to deep-water transect using sites from the CalCOFI surveys to improve our metadata.
Our ideas for this sampling opportunity started in the same way as many, literally sketched out on a piece of paper, which turned into a proposal, and has landed us here on the ship, carrying out the work. An amazing opportunity, to be sure. Our sampling plan is simple: take several discrete water samples using the CTD from each site, filter some of the water for later DNA extraction and metagenomic sequencing, concentrate cells from the remaining water for later cell-sorting and single-cell genome amplification/sequencing. Because we’re filtering large volumes of water, we’re in the wet lab. Most ships have one- a place usually near a deck where water can get spilled and it’s not the end of the world. Ours is set up for use with 142mm filter units and a small tangental flow filtration system. After filtering, everything goes in a liquid N2 dewar to keep it cold. Simple enough. For now. Post-sequencing, the workload explodes.